January 19th, 2017
Hello
Announcement: We Are Moving
- Session 5, 1/24: E62-223
- Session 6, 1/26: E51-325
- Session 7, 1/31: E62-223
- Session 8, 2/2: E51-325
Advanced Topics in Data Science
Session Titles
"Don't Write For Loops"
"Be Lazy"
"Stupid Data Frame Tricks"
"Advanced Topics in Data Science"
Gameplan
- Synthesize techniques from Sessions 2 and 3.
- Work through an example of the (data-)scientific method.
- Learn key tools for building complex data pipelines from simple building blocks.
- Impress Patrick Stewart:
The Data Science Pipeline
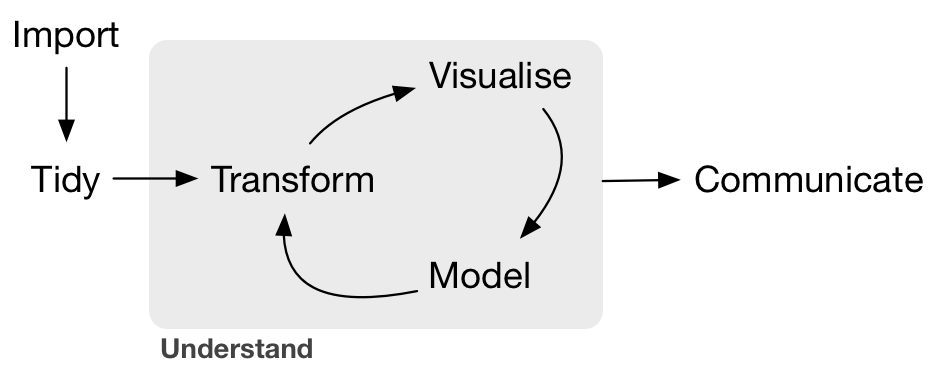 (Image credit: Hadley Wickham)
(Image credit: Hadley Wickham)
The tidyverse is a set of packages by Hadley that helps manage each stage of this pipeline.
What we've practiced already
- Import -> tidy -> transform
- Import -> tidy -> transform -> model
- Import -> tidy -> transform -> visualize
But…. 
Exercise 0
- Look left
- Look right
- Pick a partner (groups of three are fine)
You are going to need them soon.
Case Study, Part 1
- Explore the data
- Ask a question
- Form a hypothesis
What we'd like to do
Fitting a Single Model
We'd like to fit a LOESS smoother to the data to capture long-term trend. We can fit a single one like this:
model_data <- prices %>% # extract a single listing's
filter(listing_id == 5506) # worth of data
# fit a loess model, the span is a hyperparameter, a bit
# like lambda in LASSO
single_model <- loess(price_per ~ as.numeric(date),
data = model_data,
span = .5)
Examining a Single Model
single_model %>% summary()
## Call: ## loess(formula = price_per ~ as.numeric(date), data = model_data, ## span = 0.5) ## ## Number of Observations: 344 ## Equivalent Number of Parameters: 6.32 ## Residual Standard Error: 8.15 ## Trace of smoother matrix: 6.95 (exact) ## ## Control settings: ## span : 0.5 ## degree : 2 ## family : gaussian ## surface : interpolate cell = 0.2 ## normalize: TRUE ## parametric: FALSE ## drop.square: FALSE
Model Information the Tidy Way
model_preds <- broom::augment(single_model, model_data) model_preds %>% head()
## listing_id date price_per .fitted .se.fit .resid ## 1 5506 2017-09-05 72.5 73.73994 2.042531 -1.2399385 ## 2 5506 2017-09-04 72.5 73.66160 1.984215 -1.1615956 ## 3 5506 2017-09-03 72.5 73.58551 1.927151 -1.0855140 ## 4 5506 2017-09-02 72.5 73.51158 1.871376 -1.0115812 ## 5 5506 2017-09-01 72.5 73.43968 1.816931 -0.9396847 ## 6 5506 2017-08-31 72.5 73.36971 1.763855 -0.8697121
Visualizing a Single Model
model_preds %>%
ggplot(aes(x = date)) +
geom_line(aes(y = price_per)) +
geom_line(aes(y = .fitted), color = 'red')
How do we do this for multiple models?
model_container <- ????
for(id in unique(prices$listing_id)){
model <- prices %>%
filter(listing_id == id) %>%
loess(price_per ~ as.numeric(date),
data = .,
span = .25)
model_container %>% update(model) # ?????
.
.
.
}
For loops…
A familiar loop
my_list <- list('Computing', 'in', 'Optimization', 'and' ,'Statistics')
length_list <- list()
i <- 1
for(word in my_list){
length_list[i] <- nchar(word)
i <- i + 1
}
length_list %>% unlist() # just for display
## [1] 9 2 12 3 10
A better way:
my_list <- list('Computing', 'in', 'Optimization', 'and' ,'Statistics')
length_list <- map(my_list, nchar) # or my_list %>% map(nchar)
length_list %>% unlist() # just for display
## [1] 9 2 12 3 10
purrr::map() applies a function (nchar) to each entry of the original list.
Exercise
The directory exercise_data contains price data for each month. Use map to read it in efficiently, and reduce(rbind) to combine it all together. Use list.files('exercise_data', full.names = T) to get a list of all the files.
list.files('exercise_data', full.names = T) %>%
map(read_csv) %>%
reduce(rbind) %>%
head() # just for display
## # A tibble: 6 × 3 ## listing_id date price_per ## <int> <date> <dbl> ## 1 3075044 2017-04-30 32.5 ## 2 3075044 2017-04-29 37.5 ## 3 3075044 2017-04-28 37.5 ## 4 3075044 2017-04-27 32.5 ## 5 3075044 2017-04-26 32.5 ## 6 3075044 2017-04-25 32.5
Fundamental Pattern
Use lists of data frames (and data frames of lists) to organize your work.
Why?
Data frames are the fundamental unit of data science.
Usually their columns are atomic vectors of integers, doubles, dates, characters, or booleans. E.g.
## # A tibble: 6 × 3 ## listing_id date price_per ## <int> <date> <dbl> ## 1 3075044 2017-08-22 32.5 ## 2 3075044 2017-08-21 32.5 ## 3 3075044 2017-08-20 32.5 ## 4 3075044 2017-08-19 37.5 ## 5 3075044 2017-08-18 37.5 ## 6 3075044 2017-08-17 32.5
But this is somewhat inflexible. What about more complex objects? Lists can hold anything…
Well if lists are so useful…
That's a good idea!
prices_nested <- prices %>%
tidyr::nest(-listing_id)
# view the data types of the columns
map(prices_nested, class) %>% unlist()
## listing_id data ## "integer" "list"
Ew.
Actually…
prices_nested %>% head()
## # A tibble: 6 × 2 ## listing_id data ## <int> <list> ## 1 3075044 <tibble [359 × 2]> ## 2 6976 <tibble [319 × 2]> ## 3 7651065 <tibble [334 × 2]> ## 4 5706985 <tibble [344 × 2]> ## 5 2843445 <tibble [365 × 2]> ## 6 753446 <tibble [347 × 2]>
WELP
Just checking
prices_nested$data[[1]] # get the first item of the list
## # A tibble: 359 × 2 ## date price_per ## <date> <dbl> ## 1 2017-08-22 32.5 ## 2 2017-08-21 32.5 ## 3 2017-08-20 32.5 ## 4 2017-08-19 37.5 ## 5 2017-08-18 37.5 ## 6 2017-08-17 32.5 ## 7 2017-08-16 32.5 ## 8 2017-08-15 32.5 ## 9 2017-08-14 32.5 ## 10 2017-08-13 32.5 ## # ... with 349 more rows
We were here:
Now we're here:
Exercise
Use map to extract a list of lengths of each data frame in prices_nested$data
lengths <- map(prices_nested$data, nrow) lengths %>% unlist() %>% head() # for display only
## [1] 359 319 334 344 365 347
Exercise
Write a function that extracts the largest price (per person) from a data frame.
get_biggest_price <- function(data){
data$price_per %>% max(na.rm = T)
}
Now map to extract the largest price in each data frame in prices_nested$data:
biggest_prices <- map(prices_nested$data, get_biggest_price) biggest_prices %>% head() %>% unlist()
## [1] 37.50000 32.50000 39.50000 66.66667 37.50000 34.50000
Exercise
Now do the same thing, but assign the result to a new list column of prices_nested. Don't forget what you learned in Session 2!
prices_nested %>%
mutate(highest_price = map(data, get_biggest_price))
## # A tibble: 5 × 3 ## listing_id data highest_price ## <int> <list> <list> ## 1 3075044 <tibble [359 × 2]> <dbl [1]> ## 2 6976 <tibble [319 × 2]> <dbl [1]> ## 3 7651065 <tibble [334 × 2]> <dbl [1]> ## 4 5706985 <tibble [344 × 2]> <dbl [1]> ## 5 2843445 <tibble [365 × 2]> <dbl [1]>
Exercise
Now do the same thing, but assign the result to a new list column of prices_nested. Don't forget what you learned in Session 2!
prices_nested %>%
mutate(highest_price = map_dbl(data, get_biggest_price))
## # A tibble: 5 × 3 ## listing_id data highest_price ## <int> <list> <dbl> ## 1 3075044 <tibble [359 × 2]> 37.50000 ## 2 6976 <tibble [319 × 2]> 32.50000 ## 3 7651065 <tibble [334 × 2]> 39.50000 ## 4 5706985 <tibble [344 × 2]> 66.66667 ## 5 2843445 <tibble [365 × 2]> 37.50000
Summing Up
We know how to:
- Nest a data frame, creating a list of data frames.
- Use
mapto apply a function to every element of a list. - Apply a model function to an individual data frame (from Session 3).
Looks like it's time for…
Case Study, Part 2
- Fit many models
- Learn from results
- Ask new questions
We were here:
Then we did this:
Now we're here:
In code
my_loess <- function(data, span){
loess(price_per ~ as.numeric(date),
data = data,
span = span)
}
prices_nested <- prices %>%
nest(-listing_id)
prices_modeled <- prices_nested %>%
mutate(model = map(data, my_loess, span = .25))
prices_with_preds <- prices_modeled %>%
mutate(preds = map2(model, data, augment))
prices_unnested <- prices_with_preds %>%
unnest(preds)
More concise with %>%
my_loess <- function(data, span){
loess(price_per ~ as.numeric(date),
data = data,
span = span)
}
prices_unnested <- prices %>%
nest(-listing_id) %>%
mutate(model = map(data, my_loess, span = .25),
preds = map2(model, data, augment)) %>%
unnest(preds)
Even more concise
A little syntactic sugar (reference)
prices_unnested <- prices %>%
nest(-listing_id) %>%
mutate(model = map(data, ~loess(price_per ~ as.numeric(date), data = ., span = .25)),
preds = map2(model, data, augment)) %>%
unnest(preds)
Thousands of models, four lines
prices_unnested <- prices %>%
nest(-listing_id) %>%
mutate(model = map(data, ~loess(price_per ~ as.numeric(date), data = ., span = .25)),
preds = map2(model, data, augment)) %>%
unnest(preds)
Four nontrivial lines:
- Nest the data…
- …Fit a collection of the models to the data (map)…
- …Augment the data with the model predictions…
- …Unnest the data…
…and now we're ready to explore the results.
The Data Science Pipeline

Case Study Part 3
- Fit many models
- Learn from results
- Ask new questions
Finally…

Reference
Learn More About Our Packages
More on the Tidyverse
- R For Data Science is a book by Garrett Grolemund and Hadley Wickham that goes deep into a complete suite of tools for the data science pipeline.
Other "Advanced Topics in Data Science"
- Automating long data science pipelines with GNU
make. - Literate programming with RMarkdown. These slides, as well as the "pretty" lecture notes, were made in
R! - Interactive data applications with Shiny
- Develop your own
Rpackages. - Structure of the
Rlanguage – check out Advanced R.